Featured Publications
Programs, origins and immunomodulatory functions of myeloid cells in glioma
Tyler E. Miller, Chadi A. El Farran, Charles P. Couturier, Zeyu Chen, Joshua P. D’Antonio, Julia Verga, Martin A. Villanueva,L. Nicolas Gonzalez Castro, Yuzhou Evelyn Tong, Tariq Al Saadi, Andrew N. Chiocca, Yuanyuan Zhang, David S. Fischer, Dieter Henrik Heiland, Jennifer L. Guerriero, Kevin Petrecca, Mario L. Suva, Alex K. Shalek & Bradley E. Bernstein
Nature (2025)
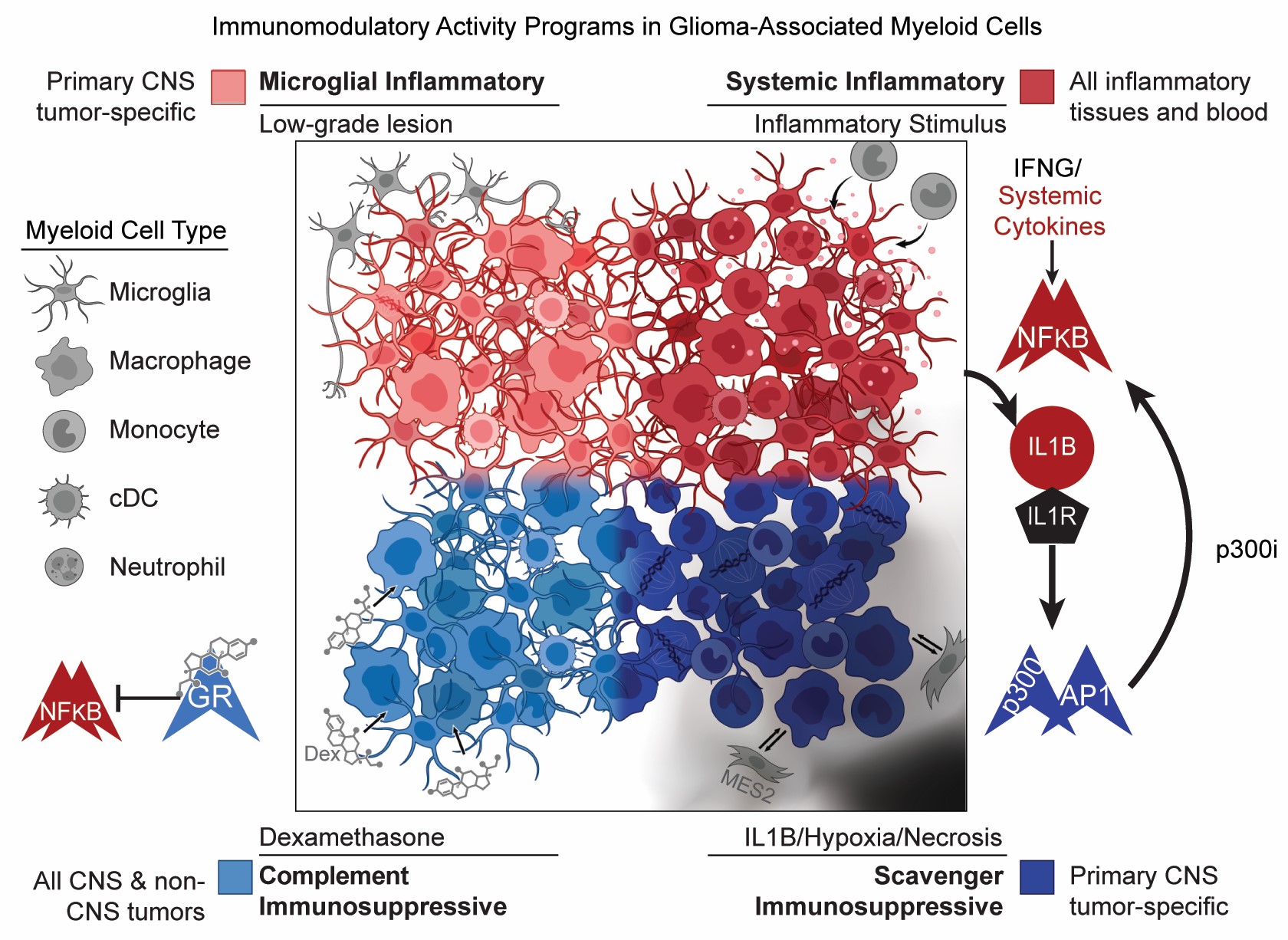
Using state of the art single cell and spatial genomic technologies, we identify novel immunosuppressive myeloid cell programs and identify ways to target them.
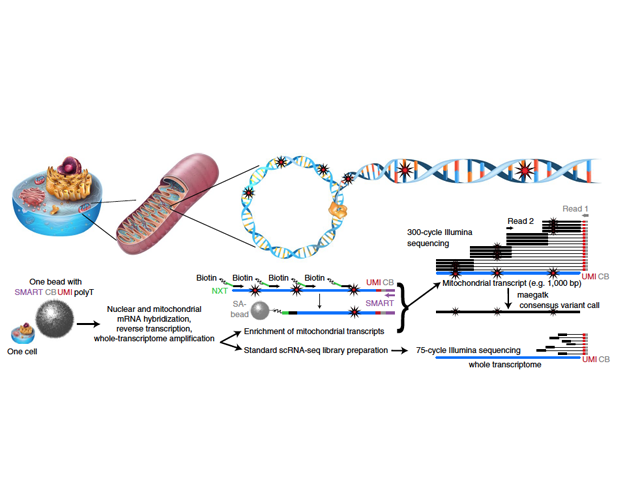
Creating single cell technology to map the origin of myeloid cells in single cells in human patients
Miller TE, El Farran CA, Couturier CP, Chen Z, D’Antonio JP, Verga J, Villanueva MA, Castro LNG, Tong YE, Saadi TA, Chiocca AN, Fischer DS, Heiland DH, Guerriero JL, Petrecca K, Suva ML, Shalek AK, Bernstein BE. Programs, Origins, and Niches of Immunomodulatory Myeloid Cells in Gliomas. bioRxiv. 2023 Oct 27;. doi: 10.1101/2023.10.24.563466. PubMed PMID: 37961527; PubMed Central PMCID: PMC10634776.
PDF
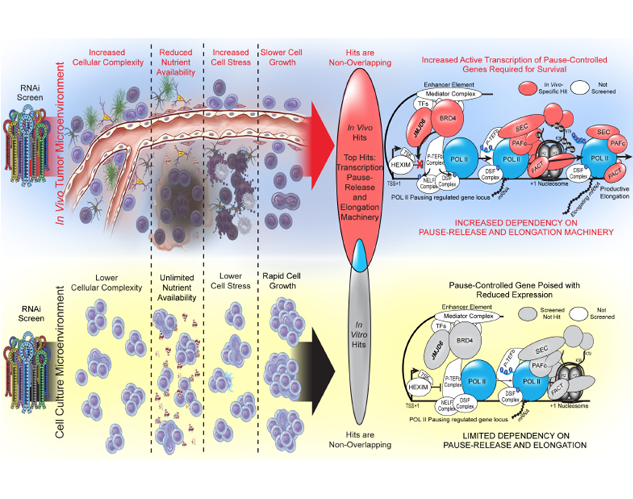
Using novel screening methods to discover novel in vivo-specific dependencies in GBM cells
Miller TE, Liau BB, Wallace LC, Morton AR, Xie Q, Dixit D, Factor DC, Kim LJY, Morrow JJ, Wu Q, Mack SC, Hubert CG, Gillespie SM, Flavahan WA, Hoffmann T, Thummalapalli R, Hemann MT, Paddison PJ, Horbinski CM, Zuber J, Scacheri PC, Bernstein BE, Tesar PJ, Rich JN. Transcription elongation factors represent in vivo cancer dependencies in glioblastoma. Nature. 2017 Jul 20;547(7663):355-359. doi: 10.1038/nature23000. Epub 2017 Jul 5. PubMed PMID: 28678782; PubMed Central PMCID: PMC5896562.
PDF
PDF to Preview in Nature
Most high-throughput target discovery screens for glioblastoma have been limited to in vitro models with uncertain physiological relevance. Here, we perform two parallel RNA interference screens for transcriptional regulators, comparing an in vitro screen in cell lines to an in vivo screen that recapitulates the tumor microenvironment. We find several transcriptional elongation factors that are specifically required for glioblastoma cell survival in vivo, particularly the transcriptional pause release factor JMJD6, which is highly expressed in gliomas. This type of in vivo functional screen has the potential to uncover novel therapeutic targets for cancer that have not been identified in previous in vitro approaches.